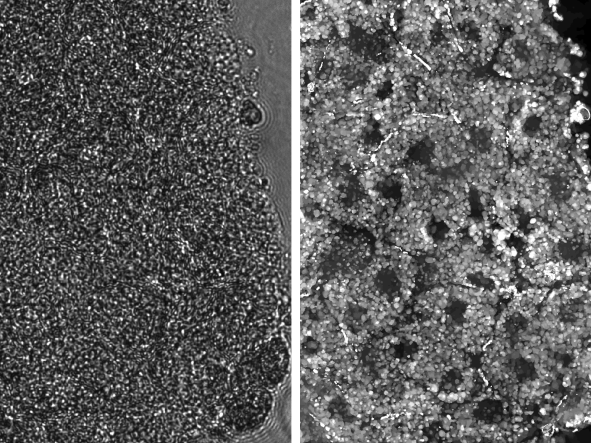
The left panel shows a raw image of Xenopus embryo tissue. This tissue is well-known to be extremely scattering, which is evident by the fact that we cannot resolve any structures in the raw image. The right panel shows the result of the new computational scatter-correction method, which drastically improves imaging capability. After scatter-correction, cellular boundaries, nuclei, and yolk platelets can be clearly identified with subcellular resolution.
Today's state-of-the-art optical imaging technologies can help us see biological dynamics occurring at subcellular resolutions. However, this capability is primarily limited to thin biological samples, such as individual cells or thin tissue sections, and falls apart when it comes to capturing high-resolution, three-dimensional images of thicker and more complex biological tissue. This limitation occurs because tissue is composed of heterogeneous arrangements of densely packed cells, which scatter light and hinder optical imaging. This is especially a challenge in live tissue, where biological dynamics occurring within the tissue further diffuse light and scuttle images.
Researchers from The University of Texas at Austin received a $1 million grant from the Chan Zuckerberg Initiative to address exactly this challenge and improve high-resolution, 3D imaging capabilities in live tissue. They plan to build a new type of imaging system that uses creative strategies to collect data. Furthermore, they plan to develop algorithms capable of unscrambling the scattered information within the data. By combining both hardware and computation, their goal is to achieve imaging capabilities that are traditionally not possible.
Why it Matters: Without the ability to get images and videos within thick live tissue, it becomes very challenging to monitor large-scale biological systems. For example, in many organisms, it is hard to fully 3D visualize internal developmental processes due to tissue scattering.
In humans, better live-imaging capabilities opens the door to improved monitoring of the brain and other vital organs and the ability to detect things as they're happening.
"This could allow you to get a more holistic view of a whole organism; it's not just two cells in a petri dish multiplying. Ideally, we want to see complex cellular interactions occurring within the organism at system-wide scales," said Shwetadwip Chowdhury, an assistant professor in the Chandra Family Department of Electrical and Computer Engineering and a co-leader on the project.
The Project: Reconstructing a 3D image typically requires hundreds of measurements. The need for all these measurements makes it nearly impossible to form a crisp and high-resolution image of tissue that is alive and moving.
The researchers will develop a next-generation optical imaging system that encodes tissue-specific information into raw measurements. They’ll also create algorithms will be developed able to use this information to get a full picture of internal tissue dynamics, even if the light from the camera becomes diffused by the live tissue, which would limit resolution in traditional image systems.
"It's kind of like trying to take a photo through a hazy, foggy environment. It's really hard to get a crisp photo in that situation, but if we design algorithms to remove the haziness, we could get a high-quality image," said Jon Tamir, an assistant professor in the Chandra Family Department of Electrical and Computer Engineering and co-leader of the study.
The Challenge: Both the hardware and software aspects of the project will prove challenging because imaging live tissue hasn't really been done before at the imaging depths that Chowdhury and Tamir are targeting. Current state-of-the-art optical technologies for deep-tissue imaging typically can see around a millimeter into tissue. For especially dense and heterogenous tissue, this limit may be even smaller. To see beyond this limit, Chowdhury and Tamir must develop an innovative imaging framework to unscramble dynamic tissue scattering. This framework will be composed of specialized strategies for data collection and space-time computational algorithms. Due to the highly complex and chaotic nature of tissue scattering, algorithms will leverage recent advances in machine learning to search for a 4D (3D space + 1D time) “solution” that best describes scattering measurements collected from the dynamic tissue sample.
It's like the old needle in the haystack metaphor, the researchers say, but without knowing what they're looking for in the haystack. The researchers say they will have to look backwards in some ways, mostly because there aren't many existing methods and prior knowledge related to what they're looking at.
"We know what individual cells look like, we know what dead tissue looks like, but we don't know what live, moving tissue looks like at long imaging depths and at high resolutions," Tamir said.
The Team: An important key to this project is the interdisciplinary nature of the team. Chowdhury's background is in developing next-generation optical imaging technologies that integrate custom hardware with computational algorithms. Chowdhury targets these technologies towards applications in science and medicine. This expertise complements that of Tamir, whose background is in magnetic resonance imaging (MRI). The challenges arising from long scan times and motion are also paramount in that field. That's why people are told to stay as still as possible when they get an MRI.
A major goal of this project is to bring together elements from both optical imaging and MRI. For example, computational approaches used in the MRI field to compensate for motion occurring within an MRI scan could potentially be adapted for optical imaging. If successful, these two fields can join together to solve a number of common problems, and the researchers hope to build a bridge between these areas of expertise and learn from each other.
"It's a great cross-pollination opportunity for us," said Chowdhury.







